Target enrichment panels with hybridization capture
Documents to download
Target enrichment panels with hybridization capture
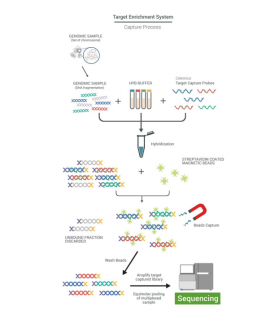
Target enrichment is a technology that allows selective sequencing of selected regions of the genome, often regions where clinically relevant nucleotide polymorphisms (mutations) are found. Compared to whole genome sequencing, it is much more time and cost efficient.
In this section we introduce products from Celemics, a South Korean company that has developed and manufactures more than 1000 target enrichment panels based on hybridization capture technology. Unlike amplicon sequencing, hybridization capture is used to detect known and unknown variants originating from distant genomic regions. Panels can contain from a few genes to all coding genes in the genome (whole-exome sequencing) and are used to detect gene variants associated with a variety of diseases and disorders. It can also accurately analyze all types of mutations such as SNV, InDel, CNV. Celemics offers complete client support including testing and rebalancing of hybridization probes. And compared to competitors, they have better coverage in GC-rich regions of the genome and sections with increased homology. Celemics' Target Enrichment Panels are compatible with samples with low quality and/or small amounts of input DNA (FFPE and cfDNA). It is possible to design a custom panel from a custom list of genes. Celemics panels can include both UMI and UDI and are compatible with most sequencing platforms and automated NGS library preparation. Also, Celemics provides a bioinformatics service and sequencing data can be uploaded to a fully EU-GDPR server where it will be processed by the Celemics bioinformatics team.
Target enrichment is used in the following areas:
Whole-exome sequencing - WES
- With a target size of 37 Mb, the Celemics WES Panel covers all clinically relevant target regions covered by the main competing WES panels on the market. The Celemics solution also includes bioinformatics support (FASTQ to Clinical Interpretation Support). Of course, there is also a Gene Add-On service that allows to add spike-in mtDNA or non-coding regions if desired.
Oncology diagnostics
- Breast/ovarian cancer - BRCA 1/2 Panel
- Hereditary cancer - OncoRisk Panel
- Somatic mutations - CancerScreen Panel (somatic cancer-related genes)
- Liquid biopsies - circulating tumor DNA (lung, colorectal, breast cancer) - ctDNA Panel
Diagnosis of inherited diseases and metabolic disorders
- G-Mendeliome clinical exome - over 7000 genes that are associated with various inherited diseases including cystic fibrosis, nephropathy, neurodegenerative disorders, cardiomyopathies, etc.
- G-Mendeliome specific panel - more suitable for smaller sequencing platforms
Pharmacogenomics
- PharmacoScreen panel - monitoring variants responsible for the success of therapies in epilepsy
Mitochondrial DNA sequencing
- Mitochondrial DNA sequencing used in research on cancer, metabolic disorders and neurological diseases associated with mtDNA - Mitochondrial DNA Sequencing Panel
RNA Sequencing
- Particularly suitable for the study of gene expression and RNA structure rearrangements - Targeted RNA Sequencing Panel
DNA methylation sequencing
- Monitoring epigenetic modifications of DNA (cytosine methylation) - Methyl-seq panel
Genotyping of viruses
- Comprehensive respiratory virus panel - for NGS genotyping of more than 30 respiratory viruses and their variants (e.g. adenoviruses, bocaviruses, coronaviruses, parainfluenza viruses, rhinoviruses, enteroviruses, etc.)
- African swine fever virus identification and genotyping panel - African Swine Fever Virus Panel
Technical details about Target enrichment panels with hybridization capture
Warranty Target enrichment panels with hybridization capture
1 Target enrichment panels with hybridization capture
2 Target enrichment panels with hybridization capture
3 Target enrichment panels with hybridization capture
4 Target enrichment panels with hybridization capture
5 Target enrichment panels with hybridization capture
6 Target enrichment panels with hybridization capture
7 Target enrichment panels with hybridization capture
8 Target enrichment panels with hybridization capture
9 Target enrichment panels with hybridization capture
10 Target enrichment panels with hybridization capture
- Producer: Celemics, Inc.

Target enrichment panels with hybridization capture
Target enrichment is a technology that allows selective sequencing of selected regions of the genome, often regions where clinically relevant nucleotide polymorphisms (mutations) are found. Compared to whole genome sequencing, it is much more time and cost efficient.
In this section we introduce products from Celemics, a South Korean company that has developed and manufactures more than 1000 target enrichment panels based on hybridization capture technology. Unlike amplicon sequencing, hybridization capture is used to detect known and unknown variants originating from distant genomic regions. Panels can contain from a few genes to all coding genes in the genome (whole-exome sequencing) and are used to detect gene variants associated with a variety of diseases and disorders. It can also accurately analyze all types of mutations such as SNV, InDel, CNV. Celemics offers complete client support including testing and rebalancing of hybridization probes. And compared to competitors, they have better coverage in GC-rich regions of the genome and sections with increased homology. Celemics' Target Enrichment Panels are compatible with samples with low quality and/or small amounts of input DNA (FFPE and cfDNA). It is possible to design a custom panel from a custom list of genes. Celemics panels can include both UMI and UDI and are compatible with most sequencing platforms and automated NGS library preparation. Also, Celemics provides a bioinformatics service and sequencing data can be uploaded to a fully EU-GDPR server where it will be processed by the Celemics bioinformatics team.
Target enrichment is used in the following areas:
Whole-exome sequencing - WES
- With a target size of 37 Mb, the Celemics WES Panel covers all clinically relevant target regions covered by the main competing WES panels on the market. The Celemics solution also includes bioinformatics support (FASTQ to Clinical Interpretation Support). Of course, there is also a Gene Add-On service that allows to add spike-in mtDNA or non-coding regions if desired.
Oncology diagnostics
- Breast/ovarian cancer - BRCA 1/2 Panel
- Hereditary cancer - OncoRisk Panel
- Somatic mutations - CancerScreen Panel (somatic cancer-related genes)
- Liquid biopsies - circulating tumor DNA (lung, colorectal, breast cancer) - ctDNA Panel
Diagnosis of inherited diseases and metabolic disorders
- G-Mendeliome clinical exome - over 7000 genes that are associated with various inherited diseases including cystic fibrosis, nephropathy, neurodegenerative disorders, cardiomyopathies, etc.
- G-Mendeliome specific panel - more suitable for smaller sequencing platforms
Pharmacogenomics
- PharmacoScreen panel - monitoring variants responsible for the success of therapies in epilepsy
Mitochondrial DNA sequencing
- Mitochondrial DNA sequencing used in research on cancer, metabolic disorders and neurological diseases associated with mtDNA - Mitochondrial DNA Sequencing Panel
RNA Sequencing
- Particularly suitable for the study of gene expression and RNA structure rearrangements - Targeted RNA Sequencing Panel
DNA methylation sequencing
- Monitoring epigenetic modifications of DNA (cytosine methylation) - Methyl-seq panel
Genotyping of viruses
- Comprehensive respiratory virus panel - for NGS genotyping of more than 30 respiratory viruses and their variants (e.g. adenoviruses, bocaviruses, coronaviruses, parainfluenza viruses, rhinoviruses, enteroviruses, etc.)
- African swine fever virus identification and genotyping panel - African Swine Fever Virus Panel
- Producer: Celemics, Inc.